Data Preparation using the Palmer Penguins Dataset
The Palmer Penguins Dataset is a popular dataset used in data science and machine learning education. The data was gathered between 2007 and 2009 by Dr. Kristen Gorman as part of the Palmer Station Long Term Ecological Research (LTER) Program. It contains measurements for 344 penguins from three species – Adelie, Chinstrap, and Gentoo – collected from three islands (Biscoe, Dream, and Torgersen) in the Palmer Archipelago, Antarctica.

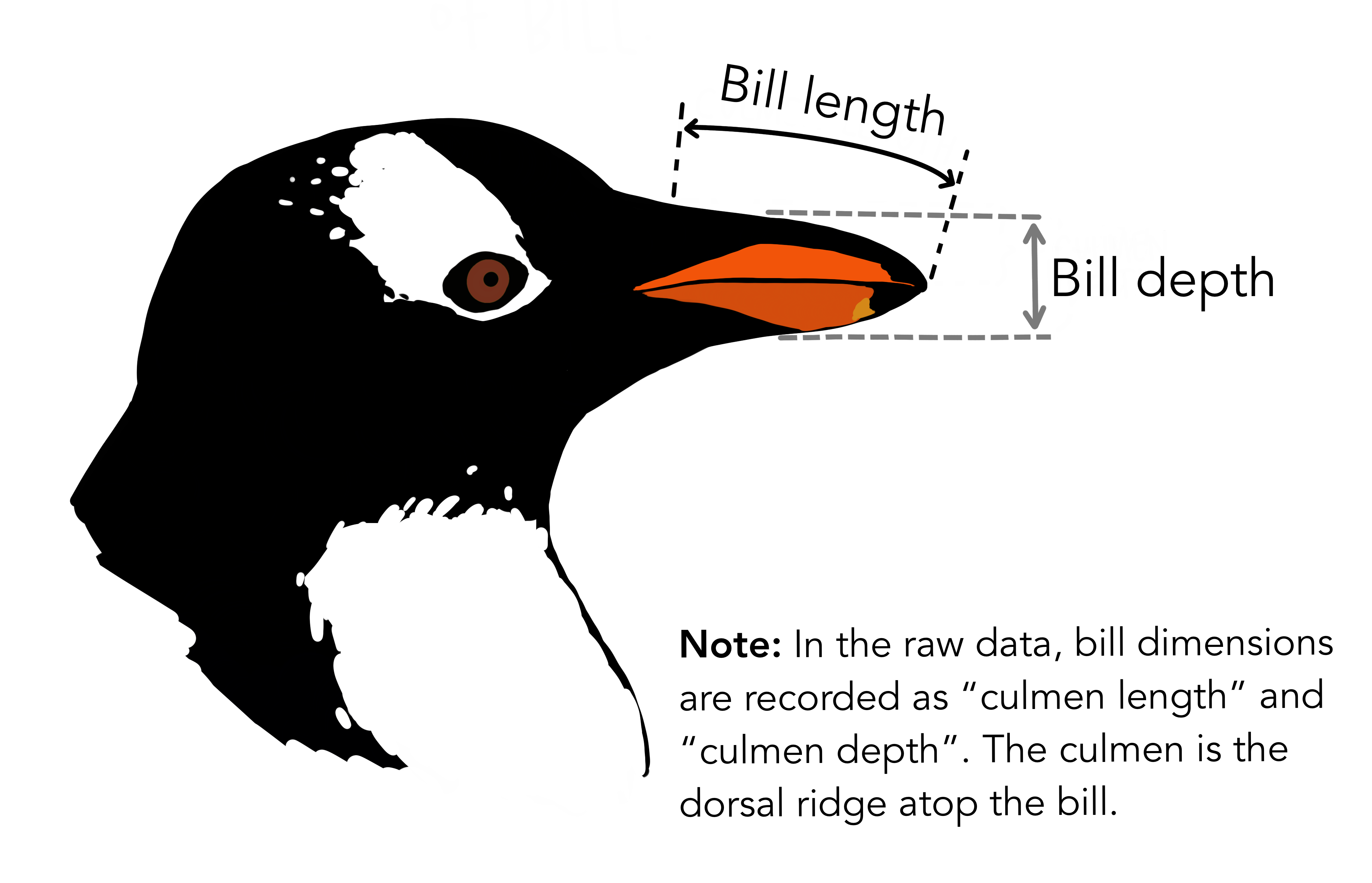
The physical attributes measured for penguins are flipper length, beak length, beak width, body mass, and sex.
1. Loading Dataset
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
# penguins = pd.read_csv("./penguins_dataset.csv")
# URL of the Penguins dataset (CSV file)
# url = "https://raw.githubusercontent.com/mwaskom/seaborn-data/master/penguins.csv"
# penguins = pd.read_csv(url)
penguins = sns.load_dataset('penguins')
penguins
penguins.info()
penguins.describe()
sns.pairplot(penguins, hue="sex", height=2.0)
sns.pairplot(penguins, hue="island", height=2.0)
sns.pairplot(penguins[["species", "bill_length_mm", "bill_depth_mm", "flipper_length_mm", "body_mass_g"]], hue="species", height=2.0)
2. Handling Missing Values
penguins_test = pd.concat([penguins.head(5), penguins.tail(5)])
penguins_test.style.highlight_null(color = 'red')
2.1 Handling missing numerical data
Mean or Median Imputation
# calculate `mean` and `median` of body_mass data
body_mass_g_mean = penguins_test.body_mass_g.mean()
body_mass_g_median = penguins_test.body_mass_g.median()
print(f" mean value of body_mass_g is {body_mass_g_mean}")
print(f"median value of body_mass_g is {body_mass_g_median}")
penguins_test['BMG_mean'] = penguins_test.body_mass_g.fillna(body_mass_g_mean)
penguins_test['BMG_median'] = penguins_test.body_mass_g.fillna(body_mass_g_median)
penguins_test.style.highlight_null(color = 'green')
plt.figure(figsize=(8, 6))
penguins_test['body_mass_g'].plot(kind='kde', color='tab:green', label="body_mass_g")
penguins_test['BMG_mean'].plot(kind='kde', color='tab:orange', label="BMG_mean")
penguins_test['BMG_median'].plot(kind='kde', color='tab:blue', label="BMG_median")
plt.legend(loc='best')
plt.tight_layout()
End of Distribution Imputation
Sometimes one would want to replace missing data with values at the tail of distribution of variable.
Advantage is that it is quick and captures importance of missing values (if one suspects missing data is valuable).
EoD’s extreme value means the mean value with 3rd stander deviation,
(mean + (3*std)))
eod_value = penguins_test['body_mass_g'].mean() + 3 * penguins_test['body_mass_g'].std()
eod_value
penguins_test['BMG_eod'] = penguins_test.body_mass_g.fillna(eod_value)
penguins_test.style.highlight_null(color = 'green')
plt.figure(figsize=(8, 6))
penguins_test['body_mass_g'].plot(kind='kde', color='tab:green', label="body_mass_g")
penguins_test['BMG_mean'].plot(kind='kde', color='tab:orange', label="BMG_mean")
penguins_test['BMG_median'].plot(kind='kde', color='tab:blue', label="BMG_median")
penguins_test['BMG_eod'].plot(kind='kde', color='tab:red', label="BMG_eod")
plt.legend(loc='best')
plt.tight_layout()
2.2 Handling missing categorical data
penguins_test.style.highlight_null(color = 'red')
# Frequent Category Imputation (mode imputation): replace missing values with the most frequent category
plt.figure(figsize=(8, 6))
penguins_test.sex.value_counts().sort_values(ascending=False).plot.bar()
plt.xlabel('Sex')
plt.ylabel('Number of Penguins')
plt.tight_layout()
print("mode for sex feature: \n", penguins_test.sex.mode())
print("Before mode imputation: \n", penguins_test['sex'], '\n')
penguins_test_sex_mode = penguins_test['sex'].fillna(penguins_test.sex.mode()[0])
print("After mode imputation: \n", penguins_test_sex_mode)
plt.figure(figsize=(8, 6))
dataset = []
female = []
male = []
dataset.append("Raw data")
female.append(penguins_test.sex.value_counts()["Female"])
male.append(penguins_test.sex.value_counts()["Male"])
dataset.append("Frequent Category Imputation")
female.append(penguins_test_sex_mode.value_counts()["Female"])
male.append(penguins_test_sex_mode.value_counts()["Male"])
x_axis = np.arange(len(dataset))
plt.bar(x_axis - 0.2, female, width=0.4, label = 'Female')
plt.bar(x_axis + 0.2, male, width=0.4, label = 'Male')
# plt.xticks(x_axis, team)
plt.xlabel('Sex')
plt.ylabel('Number of Penguins')
plt.tight_layout()
plt.legend()
Constant Value Imputation
# Constant Value Imputation
penguins_test_sex_missing = penguins_test['sex'].fillna("Missing")
print(penguins_test_sex_missing)
plt.figure(figsize=(8, 6))
penguins_test_sex_missing.value_counts().sort_values(ascending=False).plot.bar()
plt.xlabel('Sex')
plt.ylabel('Number of Penguins')
plt.tight_layout()
2.3 Remove missing values
penguins_test.style.highlight_null(color = 'red')
penguins_test_remove = penguins_test.dropna() # axis=1
penguins_test_remove.style.highlight_null(color = 'red')
3. Handling Outliers
eod_value = penguins['body_mass_g'].mean() + 3 * penguins_test['body_mass_g'].std()
print(f"EoD value = {eod_value}")
penguins_test_BMG_outlier = penguins[["species", "bill_length_mm", "body_mass_g"]]
penguins_test_BMG_outlier.loc[:, "body_mass_g"] = penguins_test_BMG_outlier["body_mass_g"].fillna(eod_value)
penguins_test_BMG_outlier
3.1 How to define outlier?
plt.figure(figsize=(8, 6))
sns.swarmplot(y=penguins_test_BMG_outlier["species"], x=penguins_test_BMG_outlier["body_mass_g"],
color="lightgreen", marker="o", size=4)
sns.boxplot(y=penguins_test_BMG_outlier["species"], x=penguins_test_BMG_outlier["body_mass_g"],
palette="coolwarm", notch=True, linewidth=2, width=0.5,
hue=penguins_test_BMG_outlier.species, legend=False)
plt.xlabel("Body mass (g)", fontsize=14)
plt.ylabel("Species", fontsize=14)
plt.tick_params(axis='x', labelsize=12)
plt.tick_params(axis='y', labelsize=12)
plt.tight_layout()
3.2 The Inter quartile range (IQR) method
penguins_test_BMG_outlier.describe()
print(f"25% quantile = {penguins_test_BMG_outlier['body_mass_g'].quantile(0.25)}")
print(f"75% quantile = {penguins_test_BMG_outlier['body_mass_g'].quantile(0.75)}\n")
IQR = penguins_test_BMG_outlier["body_mass_g"].quantile(0.75) - penguins_test_BMG_outlier["body_mass_g"].quantile(0.25)
lower_bmg_limit = penguins_test_BMG_outlier["body_mass_g"].quantile(0.25) - (1.5 * IQR)
upper_bmg_limit = penguins_test_BMG_outlier["body_mass_g"].quantile(0.75) + (1.5 * IQR)
print(f"lower limt of IQR = {lower_bmg_limit} and upper limit of IQR = {upper_bmg_limit}")
penguins_test_BMG_outlier[penguins_test_BMG_outlier["body_mass_g"] > upper_bmg_limit]
penguins_test_BMG_outlier[penguins_test_BMG_outlier["body_mass_g"] < lower_bmg_limit]
penguins_test_BMG_outlier_remove_IQR = penguins_test_BMG_outlier[penguins_test_BMG_outlier["body_mass_g"] < upper_bmg_limit]
penguins_test_BMG_outlier_remove_IQR
plt.figure(figsize=(8, 6))
sns.swarmplot(y=penguins_test_BMG_outlier_remove_IQR["species"], x=penguins_test_BMG_outlier_remove_IQR["body_mass_g"], color="lightgreen", marker="o", size=4)
sns.boxplot(y=penguins_test_BMG_outlier_remove_IQR["species"], x=penguins_test_BMG_outlier_remove_IQR["body_mass_g"],
palette="coolwarm", notch=True, linewidth=2, width=0.5,
hue=penguins_test_BMG_outlier_remove_IQR.species, legend=False)
plt.tight_layout()
3.3 The mean-standard deviation method
eod_value = penguins['body_mass_g'].mean() + 3 * penguins_test['body_mass_g'].std()
print(f"EoD value = {eod_value}")
penguins_test_BMG_outlier = penguins[["species", "bill_length_mm", "body_mass_g"]]
penguins_test_BMG_outlier.loc[:, "body_mass_g"] = penguins_test_BMG_outlier["body_mass_g"].fillna(eod_value)
penguins_test_BMG_outlier
mean = penguins_test_BMG_outlier["body_mass_g"].mean()
std = penguins_test_BMG_outlier["body_mass_g"].std()
print(f"mean = {mean}, std = {std}", '\n')
lower_bmg_limit = mean - (3.0 * std)
upper_bmg_limit = mean + (3.0 * std)
print(f"lower limt of mean-std = {lower_bmg_limit} and upper limit of mean-std = {upper_bmg_limit}")
# lower limt of IQR = 1703.125 and upper limit of IQR = 6628.125
penguins_test_BMG_outlier[penguins_test_BMG_outlier["body_mass_g"] > upper_bmg_limit]
penguins_test_BMG_outlier[penguins_test_BMG_outlier["body_mass_g"] < lower_bmg_limit]
test = penguins_test_BMG_outlier
penguins_test_BMG_outlier.loc[penguins_test_BMG_outlier["body_mass_g"] < lower_bmg_limit, "body_mass_g"] = mean
penguins_test_BMG_outlier.loc[penguins_test_BMG_outlier["body_mass_g"] > upper_bmg_limit, "body_mass_g"] = mean
penguins_test_BMG_outlier
4. Encoding Categorical Variables
penguins_sex = penguins[["species", "island", "sex"]].head(10)
penguins_sex
4.1 One hot encoding (OHE)
from sklearn.preprocessing import OneHotEncoder
encoder = OneHotEncoder(sparse_output=False) # `sparse_output=False` to get a dense array
encoded = encoder.fit_transform(penguins_sex[['sex']])
encoded = pd.DataFrame(encoded, columns=["Female", "Male", "NaN"])
penguins_sex_onehotencoding = pd.concat([penguins_sex, encoded], axis=1)
penguins_sex_onehotencoding
4.2 Label encoding
penguins_sex
from sklearn.preprocessing import LabelEncoder
encoder = LabelEncoder()
encoded = encoder.fit_transform(penguins_sex['sex'])
encoded = pd.DataFrame(encoded, columns=["sex_LE"])
penguins_sex_labelencoding = pd.concat([penguins_sex, encoded], axis=1)
penguins_sex_labelencoding
4.3 The get_dummies() function in Pandas
penguins_sex
dummy_encoded = pd.get_dummies(penguins_sex['sex']).astype(np.int8)
penguins_sex_dummyencoding = pd.concat([penguins_sex, dummy_encoded], axis=1)
penguins_sex_dummyencoding