Data science and machine learning#
Questions
Can I use Julia for machine learning?
What are the key steps in data preprocessing in Julia?
How can you handle missing data in Julia?
How can you save your current environment in Julia?
What are some popular machine learning algorithms available in Julia?
How does Julia handle large datasets in machine learning?
How can you implement clustering in Julia?
What are some classification techniques available in Julia?
Instructor note
100 min teaching
50 min exercises
Working with data#
Via Data Formats and Dataframes lesson, we explored a Julian approach to manipulating and visualization of data.
Julia is a good language to use for data science problems as it will perform well and alleviate the need to translate computationally demanding parts to another language.
Here we will learn and clustering, classification, machine learning and deep learning (toy example). Use penguin data.machine learning.
Download a dataset#
We start by downloading a dataset containing measurements of characteristic features of different penguin species.

Artwork by @allison_horst#
Todo
To obtain the data we simply add the PalmerPenguins package.
using Pkg
Pkg.add("PalmerPenguins")
using PalmerPenguins
Dataframes#
Todo
Dataframes
We will use DataFrames.jl package function here to analyze the penguins dataset, but first we need to install it:
Pkg.add("DataFrames")
using DataFrames
We now create a dataframe containing the PalmerPenguins dataset.
# using PalmerPenguins
table = PalmerPenguins.load()
df = DataFrame(table)
# the raw data can be loaded by
#tableraw = PalmerPenguins.load(; raw = true)
Summary statistics can be displayed with the describe function:
describe(df)
7×7 DataFrame
Row │ variable mean min median max nmissing eltype
│ Symbol Union… Any Union… Any Int64 Type
─────┼──────────────────────────────────────────────────────────────────────────────────────────
1 │ species Adelie Gentoo 0 String
2 │ island Biscoe Torgersen 0 String
3 │ bill_length_mm 43.9219 32.1 44.45 59.6 2 Union{Missing, Float64}
4 │ bill_depth_mm 17.1512 13.1 17.3 21.5 2 Union{Missing, Float64}
5 │ flipper_length_mm 200.915 172 197.0 231 2 Union{Missing, Int64}
6 │ body_mass_g 4201.75 2700 4050.0 6300 2 Union{Missing, Int64}
7 │ sex female male 11 Union{Missing, String}
As it was done in the Data Formats and Dataframes lesson, we can
dropmissing!(df)
The main features we are interested in for each penguin observation are bill_length_mm, bill_depth_mm, flipper_length_mm and body_mass_g. What the first three features mean is illustrated in the picture below.
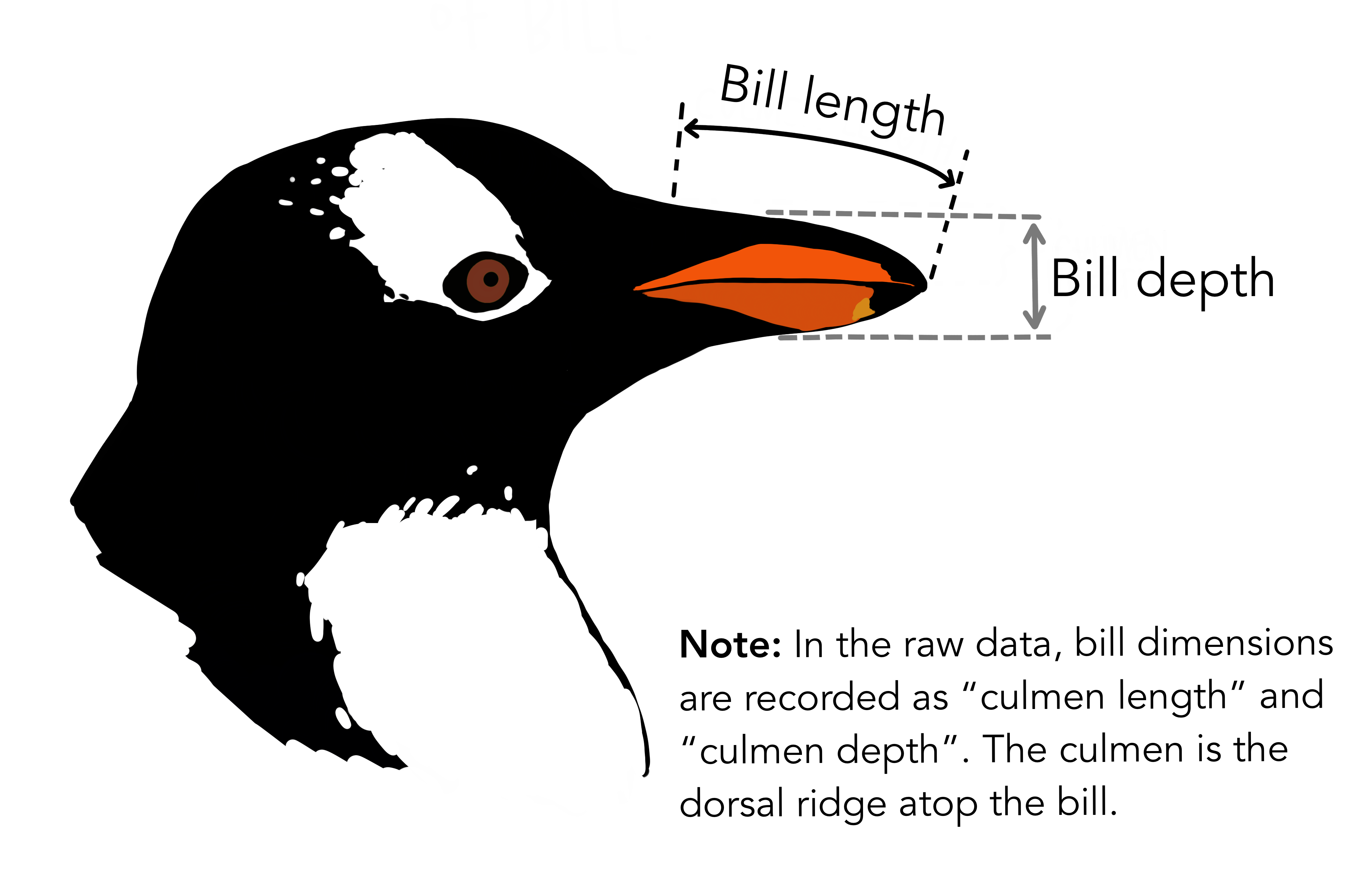
Artwork by @allison_horst#
Saving the Current Setup#
There are several ways to save the current setup in Julia. This section will cover three methods: saving the environment, saving data as a CSV file, and saving data using JLD.jl.
1. Saving the Environment#
Todo
To check the current status of your Julia environment, you can use the status command in the package manager.
using Pkg
Pkg.status()
Status `~/.julia/environments/v1.9/Project.toml`
[336ed68f] CSV v0.10.11
[aaaa29a8] Clustering v0.15.4
[a93c6f00] DataFrames v1.6.1
[682c06a0] JSON v0.21.4
[8b842266] PalmerPenguins v0.1.4
This will display the list of packages in the current environment along with their versions.
To save the state of your environment, Julia uses two files: Project.toml and Manifest.toml.
The Project.tom file specifies the packages that you explicitly added to your environment,
while the Manifest.toml file records the exact versions of these packages and all their dependencies1.
When you add packages using Pkg.add(), Julia automatically updates these files.
Therefore, your environment’s state (i.e., the set of loaded packages) is automatically saved.
Project.toml and Manifest.toml are located in the directory of your current Julia environment; in our case, ~/.julia/environments/v1.9/.
If you want to replicate this environment on another machine or in another folder, you can do the following:
Copy both
Project.tomlandManifest.tomlto the new location.In Julia, navigate to that folder and activate the environment using
Pkg.activate(".").Use
Pkg.instantiate()to download all the necessary packages.
More information in section Environments at https://enccs.github.io/julia-intro/development/
2. Saving Data as a CSV File#
(The way we use in this lesson).
Todo
(Include the content about saving data as a CSV file here)
You can use the CSV.jl package to save your DataFrame as a CSV file, which can be loaded later.
# using Pkg
# Pkg.add("CSV")
using CSV
CSV.write("penguins.csv", df)
And you can load it back with:
df = CSV.read("penguins.csv", DataFrame)
3. Saving Data Using JLD.jl#
Another option is to use JLD.jl The JLD.jl package provides a way to save and load Julia variables while preserving native types. It is a specific “dialect” of HDF5, a cross-platform, multi-language data storage format most frequently used for scientific data.
To use the JLD.jl module, you can start your code with using JLD. If you want to save a few variables and don’t care to use the more advanced features, then a simple syntax is:
using Pkg Pkg.add("JLD")Now, we can save our DataFrame df to a JLD file.
using JLD save("penguins.jld", "df", df)Here we’re saving df as “df” within penguins.jld. You can load this DataFrame back in with:
df = load("penguins.jld", "df")This will return the DataFrame df from the file and assign it back to df.
Machine learning#
Machine learning (ML) is a branch of artificial intelligence (AI) and computer science that focuses on the use of data and algorithms to imitate the way that humans learn, gradually improving its accuracy. It is an umbrella term for solving problems for which development of algorithms by human programmers would be cost-prohibitive, and instead the problems are solved by helping machines “discover” their “own” algorithms including GPT and Computer vision/Speech recognition use cases.
Now, let’s narrow our focus and look at neural networks. Neural networks (or “neural nets”, for short) are a specific choice of a model. It’s a network made up of neurons⁷. This leads to the question, “what is a neuron?” A neuron in the context of neural networks is a mathematical function conceived as a model of biological neurons. The neuron takes in one or more input values and sums them to produce an output. Normally, neurons are aggregated into layers to form a network.
For more detailed information, discover this Intro to Neurons notebook from JuliaAcademy’s Foundations of Machine Learning course. Data: draw_neural_net.jl It provides an excellent introduction to the concept of neurons in the context of ML.
References:
What is Machine Learning? – IBM. https://www.ibm.com/topics/machine-learning
Machine learning - Wikipedia. https://en.wikipedia.org/wiki/Machine_learning
1-intro-to-neurons.ipynb - Google Colab. https://colab.research.google.com/github/jigsawlabs-student/pytorch-intro-curriculum/blob/main/1-prediction-function/1-intro-to-neurons.ipynb
Machine learning in Julia#
Despite being a relatively new language, Julia already has a strong and rapidly expanding ecosystem of libraries for machine learning and deep learning. A fundamental advantage of Julia for ML is that it solves the two-language problem - there is no need for different languages for the user-facing framework and the backend heavy-lifting (like for most other DL frameworks).
A particular focus in the Julia approach to ML is “scientific machine learning” (SciML) (a.k.a. physics-informed learning), i.e. machine learning which incorporates scientific models into the learning process instead of relying only on data. The core principle of SciML is differentiable programming - the ability to automatically differentiate any code and thus incorporate it into Flux (predictive) models.
However, Julia is still behind frameworks like PyTorch and Tensorflow/Keras in terms of documentation and API design.
Traditional machine learning#
Julia has packages for traditional (non-deep) machine learning:
ScikitLearn.jl is a port of the popular Python package.
MLJ.jl provides a common interface and meta-algorithms for selecting, tuning, evaluating, composing and comparing over 150 machine learning models.
Machine Learning · Julia Packagesl: This is a website that lists various Julia packages related to machine learning, such as MLJ.jl, Knet.jl, TensorFlow.jl, DiffEqFlux.jl, FastAI.jl, ScikitLearn.jl, and many more. You can browse the packages by their popularity, alphabetical order, or update date. Each package has a brief description and a link to its GitHub repository.
AI · Julia Packages: This is another website that lists Julia packages related to artificial intelligence, such as Flux.jl, AlphaZero.jl, BrainFlow.jl, NeuralNetDiffEq.jl, Transformers.jl, MXNet.jl, and more. You can also sort the packages by different criteria and see their details.
Julia Libraries · Top Julia Machine Learning Libraries - Analytics Vidhya: This is an article that discusses some useful Julia libraries for machine learning and deep learning applications, such as computer vision and natural language processing.
We will use a few utility functions from MLJ.jl in our deep learning
exercise below, so we will need to add it to our environment:
using Pkg
Pkg.add("MLJ")
Clustering and Classification#
In this lesson, we will be exploring the use of Julia for HPDA in a Jupyter notebook environment within Visual Studio Code (VSCode).
To set up your environment, you can follow the instructions provided in the JuliaIntro lesson. This guide will walk you through the process of installing Julia, setting up JupyterLab, and adding a Julia kernel. Jupyter notebooks offer an interactive computing environment where you can combine code execution, rich text, mathematics, plots, and rich media.
Once your environment is set up, you can start using Julia in Jupyter notebooks within VSCode. This setup provides a powerful interface for writing and debugging your code. It also allows you to easily visualize your data and results.
After setting up your environment, we will dive into the adapted lessons about Clustering and Classification from the Julia MOOC on Julia Academy. These lessons provide comprehensive tutorials on various topics in Julia. By following these lessons, you will gain a deeper understanding of how to use Julia for high-performance data analysis.
Clustering notebook: ENCCS/julia-for-hpda
Classification notebook: ENCCS/julia-for-hpda
Deep learning#
Deep learning is a subset of ML which is essentially a neural network with three or more layers. These neural networks attempt to simulate the behavior of the human brain—albeit far from matching its ability—allowing it to “learn” from large amounts of data. Deep learning drives many AI applications and services that improve automation, performing analytical and physical tasks without human intervention Deep-learning architectures such as deep neural networks, deep belief networks, deep reinforcement learning, recurrent neural networks, convolutional neural networks and transformers have been applied to fields including computer vision, speech recognition, natural language processing, machine translation, bioinformatics, drug design, medical image analysis, climate science, material inspection and board game programs.
Flux.jl comes “batteries-included” with many useful tools built in, but also enables the user to write own Julia code for DL components.
Flux has relatively few explicit APIs for features like regularisation or embeddings.
All of Flux is straightforward Julia code and it can be worth to inspect and extend it if needed.
Flux works well with other Julia libraries, like dataframes, images and differential equation solvers. One can build complex data processing pipelines that integrate Flux models.
To install Flux:
using Pkg
Pkg.add("Flux")
Todo
Training a deep neural network to classify penguins
To train a model we need four things:
A collection of data points that will be provided to the objective function.
A objective (cost or loss) function, that evaluates how well a model is doing given some input data.
The definition of a model and access to its trainable parameters.
An optimiser that will update the model parameters appropriately.
First we import the required modules and load the data:
using Flux
using MLJ: partition, ConfusionMatrix
using DataFrames
using PalmerPenguins
table = PalmerPenguins.load()
df = DataFrame(table)
dropmissing!(df)
We can now preprocess our dataset to make it suitable for training a network:
# select feature and label columns
X = select(df, Not([:species, :sex, :island]))
Y = df[:, :species]
# split into training and testing parts
(xtrain, xtest), (ytrain, ytest) = partition((X, Y), 0.8, shuffle=true, rng=123, multi=true)
# use single precision and transpose arrays
xtrain, xtest = Float32.(Array(xtrain)'), Float32.(Array(xtest)')
# one-hot encoding
ytrain = Flux.onehotbatch(ytrain, ["Adelie", "Gentoo", "Chinstrap"])
ytest = Flux.onehotbatch(ytest, ["Adelie", "Gentoo", "Chinstrap"])
# count penguin classes to see if it's balanced
sum(ytrain, dims=2)
sum(ytest, dims=2)
Next up is the loss function which will be minimized during the training. We also define another function which will give us the accuracy of the model:
# we use the cross-entropy loss function typically used for classification
loss(x, y) = Flux.crossentropy(model(x), y)
# onecold (opposite to onehot) gives back the original representation
function accuracy(x, y)
return sum(Flux.onecold(model(x)) .== Flux.onecold(y)) / size(y, 2)
end
model will be our neural network, so we go ahead and define it:
n_features, n_classes, n_neurons = 4, 3, 10
model = Chain(
Dense(n_features, n_neurons, sigmoid),
Dense(n_neurons, n_classes),
softmax)
We now define an anonymous callback function to pass into the training function to monitor the progress, select the standard ADAM optimizer, and extract the parameters of the model:
callback = () -> @show(loss(xtrain, ytrain))
opt = ADAM()
θ = Flux.params(model)
Before training the model, let’s have a look at some initial predictions and the accuracy:
# predictions before training
model(xtrain[:,1:5])
ytrain[:,1:5]
# accuracy before training
accuracy(xtrain, ytrain)
accuracy(xtest, ytest)
Finally we are ready to train the model. Let’s run 100 epochs:
# the training data and the labels can be passed as tuples to train!
for i in 1:10
Flux.train!(loss, θ, [(xtrain, ytrain)], opt, cb = Flux.throttle(callback, 1))
end
# check final accuracy
accuracy(xtrain, ytrain)
accuracy(xtest, ytest)
The performance of the model is probably somewhat underwhelming, but you will fix that in an exercise below!
We finally create a confusion matrix to quantify the performance of the model:
predicted_species = Flux.onecold(model(xtest), ["Adelie", "Gentoo", "Chinstrap"])
true_species = Flux.onecold(ytest, ["Adelie", "Gentoo", "Chinstrap"])
ConfusionMatrix()(predicted_species, true_species)
Exercises#
Todo
Improve the deep learning model
Improve the performance of the neural network we trained above!
The network is not improving much because of the large numerical
range of the input features (from around 15 to around 6000) combined
with the fact that we use a sigmoid activation function. A standard
method in machine learning is to normalize features by “batch
normalization”. Replace the network definition with the following and
see if the performance improves:
n_features, n_classes, n_neurons = 4, 3, 10
model = Chain(
Dense(n_features, n_neurons),
BatchNorm(n_neurons, relu),
Dense(n_neurons, n_classes),
softmax)
Performance is usually better also if we, instead of training on the entire dataset at once, divide the training data into “minibatches” and update the network weights on each minibatch separately. First define the following function:
using StatsBase: sample
function create_minibatches(xtrain, ytrain, batch_size=32, n_batch=10)
minibatches = Tuple[]
for i in 1:n_batch
randinds = sample(1:size(xtrain, 2), batch_size)
push!(minibatches, (xtrain[:, randinds], ytrain[:,randinds]))
end
return minibatches
end
and then create the minibatches by calling the function.
You will not need to manually loop over the minibatches, simply pass
the minibatches vector of tuples to the Flux.train! function.
Does this make a difference?
Solution
function create_minibatches(xtrain, ytrain, batch_size=32, n_batch=10)
minibatches = Tuple[]
for i in 1:n_batch
randinds = sample(1:size(xtrain, 2), batch_size)
push!(minibatches, (xtrain[:, randinds], ytrain[:,randinds]))
end
return minibatches
end
n_features, n_classes, n_neurons = 4, 3, 10
model = Chain(
Dense(n_features, n_neurons),
BatchNorm(n_neurons, relu),
Dense(n_neurons, n_classes),
softmax)
callback = () -> @show(loss(xtrain, ytrain))
opt = ADAM()
θ = Flux.params(model)
minibatches = create_minibatches(xtrain, ytrain)
for i in 1:100
# train on minibatches
Flux.train!(loss, θ, minibatches, opt, cb = Flux.throttle(callback, 1));
end
accuracy(xtrain, ytrain)
# 0.9849624060150376
accuracy(xtest, ytest)
# 0.9850746268656716
predicted_species = Flux.onecold(model(xtest), ["Adelie", "Gentoo", "Chinstrap"])
true_species = Flux.onecold(ytest, ["Adelie", "Gentoo", "Chinstrap"])
ConfusionMatrix()(predicted_species, true_species)
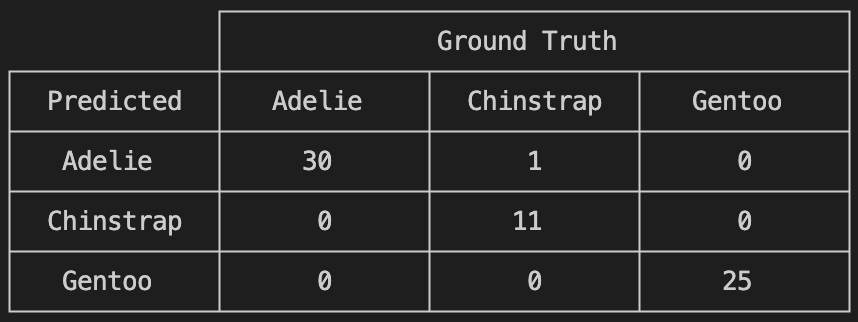
Much better!
Todo
More improvements
Exercise: Hyperparameter Tuning
Experiment with different hyperparameters of the model and the training process.
# Try different batch sizes in the minibatch creation.
minibatches = create_minibatches(xtrain, ytrain, batch_size=64, n_batch=10)
# Experiment with different learning rates for the ADAM optimizer.
opt = ADAM(0.05)
# Change the number of neurons in the hidden layer of the model.
model = Chain(
Dense(n_features, 20, relu),
Dense(20, n_classes),
softmax
)
# The solution will depend on the specific hyperparameters chosen.
Exercise: Feature Engineering
Consider doing some feature engineering on your input data.
# Try normalizing or standardizing the input features.
xtrain = (xtrain .- mean(xtrain, dims=2)) ./ std(xtrain, dims=2)
xtest = (xtest .- mean(xtest, dims=2)) ./ std(xtest, dims=2)
Exercise: Different Model Architectures
Experiment with different model architectures.
# Try adding more layers to your model.
model = Chain(
Dense(n_features, n_neurons, relu),
Dense(n_neurons, n_neurons, relu),
Dense(n_neurons, n_classes),
softmax
)
Remember to experiment and see how these changes affect your model’s performance! 😊
See also#
Many interesting datasets are available in Julia through the RDatasets package. For instance:
Pkg.add("RDatasets") using RDatasets # load a couple of datasets iris = dataset("datasets", "iris") neuro = dataset("boot", "neuro")
“The Future of Machine Learning and why it looks a lot like Julia” by Logan Kilpatrick
Neuromorphic | Probabilistic learning#
Nordic Neuromorphs | NorN Discord Community – https://discord.gg/5Qq6yX5
Quantum#
Swedish Quantum Society | SQS – https://swedishquantumsociety.vercel.app/
