Performance boosting
Objectives
Learn how to boost performance using Numba and Cython
Instructor note
20 min teaching/type-along
20 min exercises
After benchmarking and optimizing your code, you can start thinking of accelerating it further with libraries like Cython and Numba to pre-compile performance-critical functions.
Pre-compiling Python
For many (or most) use cases, using NumPy or Pandas is sufficient. However, in some computationally heavy applications, it is possible to improve the performance by pre-compiling expensive functions. Cython and Numba are among the popular choices and both of them have good support for NumPy arrays.
Cython
Cython is a superset of Python that additionally supports calling C functions and declaring C types on variables and class attributes. Under Cython, source code gets translated into optimized C/C++ code and compiled as Python extension modules.
Developers can run the cython command-line utility to produce a .c file from
a .py file which needs to be compiled with a C compiler to an .so library
which can then be directly imported in a Python program. There is, however, also an easy
way to use Cython directly from Jupyter notebooks through the %%cython magic
command. We will restrict the discussion here to the Jupyter-way. For a full overview
of the capabilities refer to the documentation.
Demo: Cython
Consider the following pure Python code which integrates a function:
import numpy as np
def f(x):
return x ** 2 - x
def integrate_f(a, b, N):
s = 0
dx = (b - a) / N
for i in range(N):
s += f(a + i * dx)
return s * dx
def apply_integrate_f(col_a, col_b, col_N):
n = len(col_N)
res = np.empty(n,dtype=np.float64)
for i in range(n):
res[i] = integrate_f(col_a[i], col_b[i], col_N[i])
return res
We generate a dataframe and apply the apply_integrate_f() function on its columns, timing the execution:
import pandas as pd
df = pd.DataFrame({"a": np.random.randn(1000),
"b": np.random.randn(1000),
"N": np.random.randint(100, 1000, (1000))})
%timeit apply_integrate_f(df['a'], df['b'], df['N'])
# 321 ms ± 10.7 ms per loop (mean ± std. dev. of 7 runs, 1 loop each)
In order to use Cython, we need to import the Cython extension:
%load_ext cython
As a first cythonization step we add the cython magic command with the
-a, --annotate flag, %%cython -a, to the top of the Jupyter code cell.
The yellow coloring in the output shows us the amount of pure Python:

Our task is to remove as much yellow as possible by static typing, i.e. explicitly declaring arguments, parameters, variables and functions. We can start by simply compiling the code using Cython without any changes:
%%cython
import numpy as np
import pandas as pd
def f_cython(x):
return x * (x - 1)
def integrate_f_cython(a, b, N):
s = 0
dx = (b - a) / N
for i in range(N):
s += f_cython(a + i * dx)
return s * dx
def apply_integrate_f_cython(col_a, col_b, col_N):
n = len(col_N)
res = np.empty(n,dtype=np.float64)
for i in range(n):
res[i] = integrate_f_cython(col_a[i], col_b[i], col_N[i])
return res
%timeit apply_integrate_f_cython(df['a'], df['b'], df['N'])
# 276 ms ± 20.2 ms per loop (mean ± std. dev. of 7 runs, 1 loop each)
Simply by using Cython and a copy-and-paste gives us about 10% increase in performance.
Now we can start adding data type annotation to the input variables:
%%cython
import numpy as np
import pandas as pd
def f_cython_dtype0(double x):
return x ** 2 - x
def integrate_f_cython_dtype0(double a, double b, long N):
s = 0
dx = (b - a) / N
for i in range(N):
s += f_cython_dtype0(a + i * dx)
return s * dx
def apply_integrate_f_cython_dtype0(double[:] col_a, double[:] col_b, long[:] col_N):
n = len(col_N)
res = np.empty(n,dtype=np.float64)
for i in range(n):
res[i] = integrate_f_cython_dtype0(col_a[i], col_b[i], col_N[i])
return res
# this will not work
#%timeit apply_integrate_f_cython_dtype0(df['a'], df['b'], df['N'])
# but rather
%timeit apply_integrate_f_cython_dtype0(df['a'].to_numpy(), df['b'].to_numpy(), df['N'].to_numpy())
# 41.4 ms ± 1.27 ms per loop (mean ± std. dev. of 7 runs, 10 loops each)
Warning
You can not pass a Series directly since the Cython definition is specific to an array.
Instead using the Series.to_numpy() to get the underlying NumPy array
which works nicely with Cython.
Cython uses the normal C syntax for types and provides all standard ones, including pointers. Here is a list of a few examples:
NumPy dtype
Cython type identifier
C type identifier
import numpy as np
cimport numpy as cnp
N/A
N/A
cnp.int_t
long
np.intc
N/A
int
np.intp
cnp.intp_t
ssize_t
np.int8
cnp.int8_t
signed char
np.int16
cnp.int16_t
signed short
np.int32
cnp.int32_t
signed int
np.int64
cnp.int64_t
signed long long
np.uint8
cnp.uint8_t
unsigned char
np.uint16
cnp.uint16_t
unsigned short
np.uint32
cnp.uint32_t
unsigned int
np.uint64
cnp.uint64_t
unsigned long
cnp.float64_t
double
np.float32
cnp.float32_t
float
np.float64
cnp.float64_t
double
cnp.complex128_t
double complex
np.complex64
cnp.complex64_t
float complex
np.complex128
cnp.complex128_t
double complex
Differeces between cimport and import statements
cimport gives access to C functions or attributes
import gives access to Python functions or attributes
it is common to use the following, and Cython will internally handle this ambiguity
import numpy as np # access to NumPy Python functions cimport numpy as np # access to NumPy C API
Next step, we can start adding type annotation to the functions. There are three ways of declaring functions:
def- Python style:
Called by Python or Cython code, and both input/output are Python objects.
Declaring the types of arguments and local types (thus return values) can allow Cython
to generate optimized code which speeds up the execution. Once the types are declared,
a TypeError will be raised if the function is passed with the wrong types.
cdef- C style:
Called from Cython and C, but not from Python code.
Cython treats the function as pure C functions, which can take any type of arguments,
including non-Python types, e.g. pointers. It will give you the best performance.
However, one should really take care of the cdef declared functions,
since you are actually writing in C.
cpdef- Python/C mixed:
cpdef function combines both def and cdef. Cython will generate a cdef
function for C types and a def function for Python types. In terms of performance,
cpdef functions may be as fast as those using cdef and might be as slow as
def declared functions.
%%cython
import numpy as np
import pandas as pd
cdef f_cython_dtype1(double x):
return x ** 2 - x
cpdef integrate_f_cython_dtype1(double a, double b, long N):
s = 0
dx = (b - a) / N
for i in range(N):
s += f_cython_dtype1(a + i * dx)
return s * dx
cpdef apply_integrate_f_cython_dtype1(double[:] col_a, double[:] col_b, long[:] col_N):
n = len(col_N)
res = np.empty(n,dtype=np.float64)
for i in range(n):
res[i] = integrate_f_cython_dtype1(col_a[i], col_b[i], col_N[i])
return res
%timeit apply_integrate_f_cython_dtype1(df['a'].to_numpy(), df['b'].to_numpy(), df['N'].to_numpy())
# 37.2 ms ± 556 µs per loop (mean ± std. dev. of 7 runs, 10 loops each)
Last step, we can add type annotation to the local variables within the functions and the output.
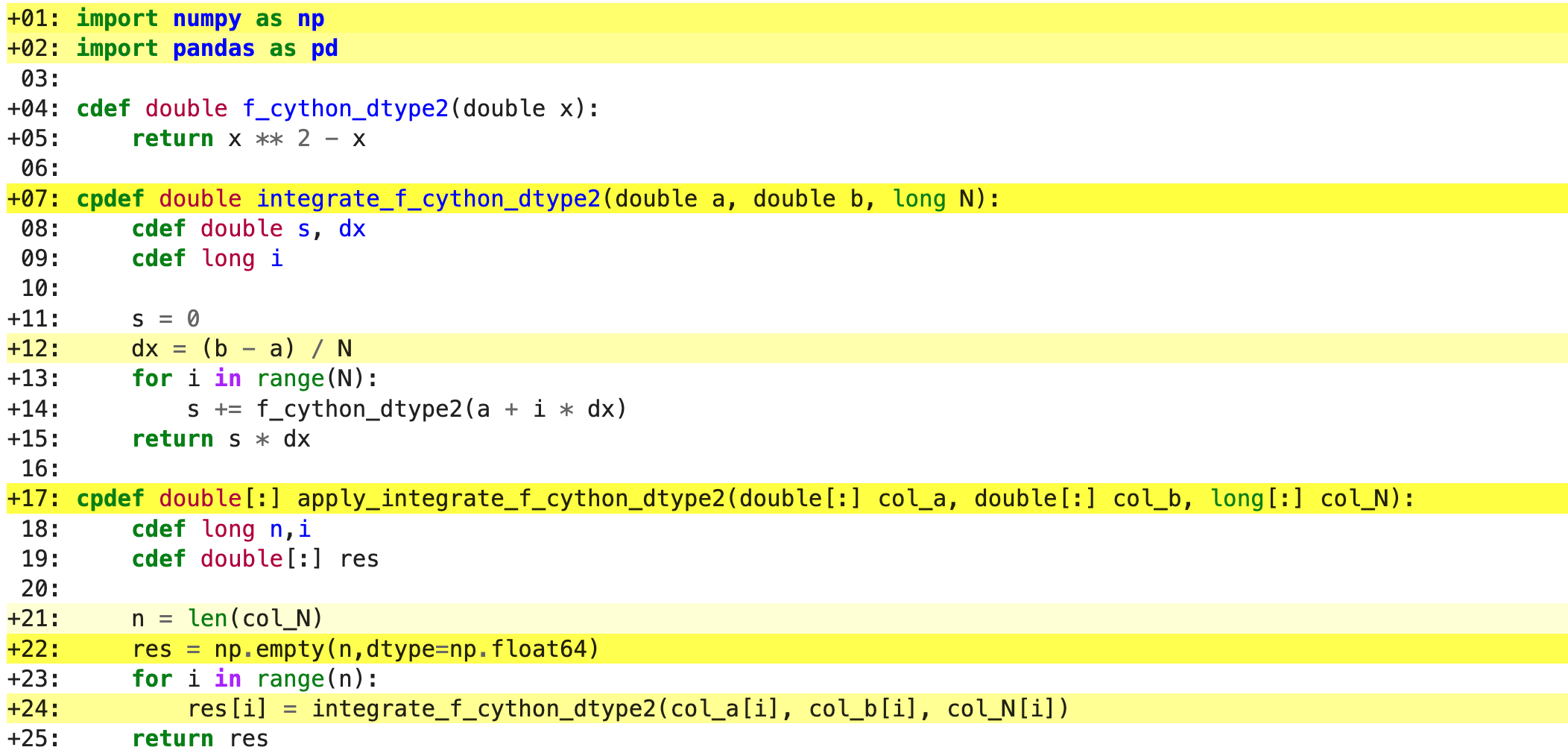
%%cython
import numpy as np
import pandas as pd
cdef double f_cython_dtype2(double x):
return x ** 2 - x
cpdef double integrate_f_cython_dtype2(double a, double b, long N):
cdef double s, dx
cdef long i
s = 0
dx = (b - a) / N
for i in range(N):
s += f_cython_dtype2(a + i * dx)
return s * dx
cpdef double[:] apply_integrate_f_cython_dtype2(double[:] col_a, double[:] col_b, long[:] col_N):
cdef long n,i
cdef double[:] res
n = len(col_N)
res = np.empty(n,dtype=np.float64)
for i in range(n):
res[i] = integrate_f_cython_dtype2(col_a[i], col_b[i], col_N[i])
return res
%timeit apply_integrate_f_cython_dtype2(df['a'].to_numpy(), df['b'].to_numpy(), df['N'].to_numpy())
# 696 µs ± 8.71 µs per loop (mean ± std. dev. of 7 runs, 1,000 loops each)
Now it is over 400 times faster than the original Python implementation, and all we have done is to add
type declarations! If we add the -a annotation flag we indeed see much less Python interaction in the
code.
Numba
An alternative to statically compiling Cython code is to use a dynamic just-in-time (JIT) compiler with Numba.
Numba allows you to write a pure Python function which can be JIT compiled to native machine instructions,
similar in performance to C, C++ and Fortran, by simply adding the decorator @jit in your function.
However, the @jit compilation will add overhead to the runtime of the function,
i.e. the first time a function is run using Numba engine will be slow as Numba will have the function compiled.
Once the function is JIT compiled and cached, subsequent calls will be fast. So the performance benefits may not be
realized especially when using small datasets.
Numba supports compilation of Python to run on either CPU or GPU hardware and is designed to integrate with the Python scientific software stack. The optimized machine code is generated by the LLVM compiler infrastructure.
Demo: Numba
Consider the integration example again using Numba this time:
import numpy as np
import numba
@numba.jit
def f_numba(x):
return x ** 2 - x
@numba.jit
def integrate_f_numba(a, b, N):
s = 0
dx = (b - a) / N
for i in range(N):
s += f_numba(a + i * dx)
return s * dx
@numba.jit
def apply_integrate_f_numba(col_a, col_b, col_N):
n = len(col_N)
res = np.empty(n,dtype=np.float64)
for i in range(n):
res[i] = integrate_f_numba(col_a[i], col_b[i], col_N[i])
return res
# try passing Pandas Series
%timeit apply_integrate_f_numba(df['a'],df['b'],df['N'])
# 6.02 ms ± 56.5 µs per loop (mean ± std. dev. of 7 runs, 1 loop each)
# try passing NumPy array
%timeit apply_integrate_f_numba(df['a'].to_numpy(),df['b'].to_numpy(),df['N'].to_numpy())
# 625 µs ± 697 ns per loop (mean ± std. dev. of 7 runs, 1,000 loops each)
Warning
Numba is best at accelerating functions that apply numerical functions to NumPy arrays. When used with Pandas,
pass the underlying NumPy array of Series or DataFrame (using to_numpy()) into the function.
If you try to @jit a function that contains unsupported Python or NumPy code, compilation will fall back to the object mode
which will mostly likely be very slow. If you would prefer that Numba throw an error for such a case,
you can do e.g. @numba.jit(nopython=True) or @numba.njit.
We can further add date type, although in this case there is not much performance improvement:
import numpy as np
import numba
@numba.jit(numba.float64(numba.float64))
def f_numba_dtype(x):
return x ** 2 - x
@numba.jit(numba.float64(numba.float64,numba.float64,numba.int64))
def integrate_f_numba_dtype(a, b, N):
s = 0
dx = (b - a) / N
for i in range(N):
s += f_numba_dtype(a + i * dx)
return s * dx
@numba.jit(numba.float64[:](numba.float64[:],numba.float64[:],numba.int64[:]))
def apply_integrate_f_numba_dtype(col_a, col_b, col_N):
n = len(col_N)
res = np.empty(n,dtype=np.float64)
for i in range(n):
res[i] = integrate_f_numba_dtype(col_a[i], col_b[i], col_N[i])
return res
%timeit apply_integrate_f_numba_dtype(df['a'].to_numpy(),df['b'].to_numpy(),df['N'].to_numpy())
# 625 µs ± 697 ns per loop (mean ± std. dev. of 7 runs, 1,000 loops each)
Numba vs Cython
Should you use Numba or Cython? Does it matter?
Performance is usually very similar and exact results depend on versions of Python, Cython, Numba and NumPy.
Numba is generally easier to use (just add
@jit)Cython is more stable and mature, Numba developing faster
Numba also works for GPUs
Cython can compile arbitrary Python code and directly call C libraries, Numba has restrictions
Numba requires LLVM toolchain, Cython only C compiler.
Finally:
NumPy is really good at what it does. For simple operations or small data, Numba or Cython is not going to outperform it. But when things get more complex these frameworks can save the day!
Exercises
Profile the word-autocorrelation code
Revisit the word-autocorrelation code. To clone the repository (if you haven’t already):
$ git clone https://github.com/ENCCS/word-count-hpda.git
To run the code, type:
$ cd word-count-hpda
$ python source/wordcount.py data/pg99.txt processed_data/pg99.dat
$ python source/autocorrelation.py data/pg99.txt processed_data/pg99.dat results/acf_pg99.dat
Add @profile to the word_acf() function, and run kernprof
from the command line. What lines of this function are the most expensive?
Solution
$ kernprof -l -v source/autocorrelation.py data/pg99.txt processed_data/pg99.dat results/acf_pg99.dat
Output:
Wrote profile results to autocorrelation.py.lprof
Timer unit: 1e-06 s
Total time: 15.5976 s
File: source/autocorrelation.py
Function: word_acf at line 24
Line # Hits Time Per Hit % Time Line Contents
==============================================================
24 @profile
25 def word_acf(word, text, timesteps):
26 """
27 Calculate word-autocorrelation function for given word
28 in a text. Each word in the text corresponds to one "timestep".
29 """
30 10 1190.0 119.0 0.0 acf = np.zeros((timesteps,))
31 10 15722.0 1572.2 0.1 mask = [w==word for w in text]
32 10 6072.0 607.2 0.0 nwords_chosen = np.sum(mask)
33 10 14.0 1.4 0.0 nwords_total = len(text)
34 1010 658.0 0.7 0.0 for t in range(timesteps):
35 11373500 4675124.0 0.4 30.0 for i in range(1,nwords_total-t):
36 11372500 10897305.0 1.0 69.9 acf[t] += mask[i]*mask[i+t]
37 1000 1542.0 1.5 0.0 acf[t] /= nwords_chosen
38 10 10.0 1.0 0.0 return acf
Is the word_acf() function efficient?
Have another look at the word_acf() function from the word-count project.
def word_acf(word, text, timesteps):
"""
Calculate word-autocorrelation function for given word
in a text. Each word in the text corresponds to one "timestep".
"""
acf = np.zeros((timesteps,))
mask = [w==word for w in text]
nwords_chosen = np.sum(mask)
nwords_total = len(text)
for t in range(timesteps):
for i in range(1,nwords_total-t):
acf[t] += mask[i]*mask[i+t]
acf[t] /= nwords_chosen
return acf
Do you think there is any room for improvement? How would you go about optimizing this function?
Try to implement one faster version!
Hints
You can replace the double loop (the manual calculation of an ACF) with an in-built NumPy function,
np.correlate(). NumPy gurus often know which function to use for which algorithms, but searching the internet also helps. One typically needs to figure out how to use the in-built function for the particular use case.There are two ways of using Numba, one with
nopython=Falseand one withnopython=True. The latter needs a rewrite of theword_acf()function to accept themaskarray, since Numba cannot pre-compile the expression definingmask.
Solution
The function uses a Python object (mask) inside a double for-loop,
which is guaranteed to be suboptimal. There are a number of ways to speed
it up. One is to use numba and just-in-time compilation, as we shall
see below.
Another is to find an in-built vectorized NumPy function which can calculate the
autocorrelation for us! Here are the Numpy and Numba (nopython=False) versions:
def word_acf_numpy(word, text, timesteps):
"""
Calculate word-autocorrelation function for given word
in a text using numpy.correlate function.
Each word in the text corresponds to one "timestep".
"""
mask = np.array([w==word for w in text]).astype(np.float64)
acf = np.correlate(mask, mask, mode='full') / np.sum(mask)
return acf[int(acf.size/2):int(acf.size/2)+timesteps]
@numba.jit(nopython=False, cache=True)
def word_acf_numba_py(word, text, timesteps):
"""
Calculate word-autocorrelation function for given word
in a text. Each word in the text corresponds to one "timestep".
"""
acf = np.zeros((timesteps,))
mask = np.array([w==word for w in text]).astype(np.float64)
nwords_chosen = np.sum(mask)
nwords_total = len(text)
for t in range(timesteps):
for i in range(1,nwords_total-t):
acf[t] += mask[i]*mask[i+t]
acf[t] /= nwords_chosen
return acf
In the autocorr-numba-numpy branch
of the word-count-hpda repository you
can additionally find a nopython=True Numba version as well as benchmarking
of all the versions. Note that the Numba functions use cache=True to save the
precompiled code so that subsequent executions of the autocorrelation.py script
are faster than the first.
Pairwise distance
Consider the following Python function:
import numpy as np
def dis_python(X):
M = X.shape[0]
N = X.shape[1]
D = np.empty((M, M), dtype=np.float64)
for i in range(M):
for j in range(M):
d = 0.0
for k in range(N):
tmp = X[i, k] - X[j, k]
d += tmp * tmp
D[i, j] = np.sqrt(d)
return D
Start by profiling it in Jupyter:
X = np.random.random((1000, 3))
%timeit dis_python(X)
Now try to speed it up with NumPy (i.e. vectorise the function), Numba or Cython (depending on what you find most interesting).
Solution
import numpy as np
def dis_numpy(X):
return np.sqrt(((X[:, None, :] - X) ** 2).sum(-1))
X = np.random.random((1000, 3))
%timeit dis_numpy(X)
%%cython
import numpy as np
def dis_cython(double[:,:] X):
cdef ssize_t i,j,k,M,N
cdef double d,tmp
cdef double[:,:] D
M = X.shape[0]
N = X.shape[1]
D = np.empty((M, M), dtype=np.float64)
for i in range(M):
for j in range(M):
d = 0.0
for k in range(N):
tmp = X[i, k] - X[j, k]
d += tmp * tmp
D[i, j] = np.sqrt(d)
return D
X = np.random.random((1000, 3))
%timeit dis_cython(X)
We can further improve performance by using more C functions:
%%cython
import numpy as np
from libc.math cimport sqrt
def dis_cython_v1(double[:,:] X):
cdef ssize_t i,j,k,M,N
cdef double d,tmp
cdef double[:,:] D
M = X.shape[0]
N = X.shape[1]
D = np.empty((M, M), dtype=np.float64)
for i in range(M):
for j in range(M):
d = 0.0
for k in range(N):
tmp = X[i, k] - X[j, k]
d += tmp * tmp
D[i, j] = sqrt(d)
return D
X = np.random.random((1000, 3))
%timeit dis_cython_v1(X)
import numpy as np
import numba
@numba.jit
def dis_numba(X):
M = X.shape[0]
N = X.shape[1]
D = np.empty((M, M), dtype=np.float64)
for i in range(M):
for j in range(M):
d = 0.0
for k in range(N):
tmp = X[i, k] - X[j, k]
d += tmp * tmp
D[i, j] = np.sqrt(d)
return D
X = np.random.random((1000, 3))
%timeit dis_numba(X)
from scipy.spatial.distance import cdist
X = np.random.random((1000, 3))
%timeit cdist(X, X)
Bubble sort
To make a long story short, in the worse case the time taken by the Bubblesort algorithm is roughly \(O(n^2)\) where \(n\) is the number of items being sorted.
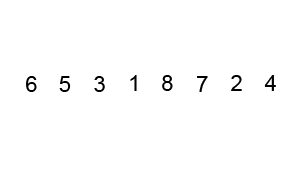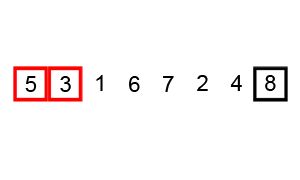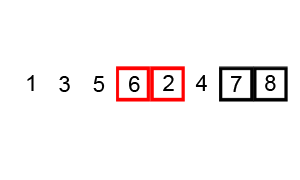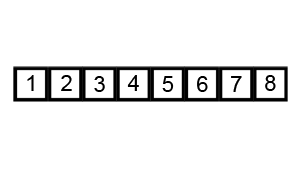
Here is a function that performs bubble-sort:
def bs_python(a_list):
N = len(a_list)
for i in range(N):
for j in range(1, N-i):
if a_list[j] < a_list[j-1]:
a_list[j-1], a_list[j] = a_list[j], a_list[j-1]
return a_list
And this is how you can benchmark it:
import random
l = [random.randint(1,1000) for num in range(1, 1000)]
%timeit bs_python(l)
Now try to speed it up with Numba or Cython (depending on what you find
most interesting). Make sure that you’re getting the correct result,
and then benchmark it with %timeit.
Solution
%%cython
cpdef long[:] bs_cython(long[:] a_list):
cdef int N, i, j
N = len(a_list)
for i in range(N):
for j in range(1, N-i):
if a_list[j] < a_list[j-1]:
a_list[j-1], a_list[j] = a_list[j], a_list[j-1]
return a_list
import random
import numpy as np
l = [random.randint(1,1000) for num in range(1, 1000)]
l_arr = np.asarray(l)
%timeit bs_cython(l_arr)
We can further improve performance by using more C/C++ features:
%%cython
cimport cython
from libc.stdlib cimport malloc, free
cpdef bs_clist(a_list):
cdef int *c_list
c_list = <int *>malloc(len(a_list)*cython.sizeof(int))
cdef int N, i, j
N = len(a_list)
for i in range(N):
c_list[i] = a_list[i]
for i in range(N):
for j in range(1, N-i):
if c_list[j] < c_list[j-1]:
c_list[j-1], c_list[j] = c_list[j], c_list[j-1]
for i in range(N):
a_list[i] = c_list[i]
free(c_list)
return a_list
import random
l = [random.randint(1,1000) for num in range(1, 1000)]
%timeit bs_clist(l)
import numba
@numba.jit
def bs_numba(a_list):
N = len(a_list)
for i in range(N):
for j in range(1, N-i):
if a_list[j] < a_list[j-1]:
a_list[j-1], a_list[j] = a_list[j], a_list[j-1]
return a_list
import random
import numpy as np
l = [random.randint(1,1000) for num in range(1, 1000)]
# first try using a list as input
%timeit bs_numba(l)
# try using a NumPy array
l = [random.randint(1,1000) for num in range(1, 1000)]
l_arr = np.asarray(l)
%timeit bs_numba(l_arr)
Static typing
Consider the following example of calculating the squre of an array. We have a few different versions using Numba. Benchmark them and compare the results.
import numpy as np X=np.random.rand(10000) %timeit np.square(X)import numpy as np import numba @numba.jit def nb_no_typing(arr): res=np.empty(len(arr)) for i in range(len(arr)): res[i]=arr[i]**2 return resX=np.random.rand(10000) %timeit nb_no_typing(X)import numpy as np import numba @numba.jit(numba.float64[:](numba.float64[:])) def nb_typing(arr): res=np.empty(len(arr)) for i in range(len(arr)): res[i]=arr[i]**2 return resX=np.random.rand(10000) %timeit nb_typing(X)import numpy as np import numba @numba.jit(numba.float64[::1](numba.float64[::1])) def nb_typing_vec(arr): res=np.empty(len(arr)) for i in range(len(arr)): res[i]=arr[i]**2 return resX=np.random.rand(10000) %timeit nb_typing_vec(X)
Typing is not a necessity
None of the Numba version will outperform the NumPy version for this simple and small example.
You will see that using static typing actually leads to a decreased performance, since the input array is not assumed to be continuous and therefore ruling out the vectorization. By defining the array as continuous, the performace can be recovered.
In principle, such cases where typing does not allow optimizations could happen to Cython codes as well, so one should always optimize where and when needed.
Keypoints
To squeeze the last drop of performance out of your Python code you can convert performance-critical functions to Numba or Cython
Both Numba and Cython pre-compile Python code to make it run faster.